R Notes
Page Contents
References \ Reads
- R Language Definition.
- The Art Of R Programming.
- Producing Simple Graphs with R, by Frank McCown
Some Basic Command Line Misc Stuff
Getting Help
To get help on a function you can use the following...
?func-name ## Display help for function with this exact name
help.search("func name") ## Search all help for function like this
args("func name") ## gets args for this exact function
Typing a function name without the ending curley brackets displays the code for the function.
Working Directory
Functions will generally read or write to your working directory (unless you use absolute paths)
Use getwd() to display your current working directory and
setwd("...") to set your current working directory.
dir("...") will list a directory.
Source R code from console
Use source("..filename..").
Basic Flow Control
If-then-else
If-the-else as standard...
if(...) {
} else if {
} else {
}
You can also assign from an if into a variable...
> x <- if(FALSE) { cos(22) } else { sin(22) }
> x
[1] -0.008851309
> x <- if(TRUE) { cos(22) } else { sin(22) }
> x
[1] -0.9999608
For loops
For each value in a range...
for(i in 1:10) { ... }
To generate an index for each member of a vector...
for(i in seq_along(x)) {
...x[i]...
}
For each item in a list...
for(item in x) { ... }
While loops
count <- 0
while(count < 10) {
count <- count + 1
}
Infinite loops
repeat {
// infinite loop
}
Skip loop iteration or exit loop
The standard break statement, and to continue use next
Functions
The basics...
In R, functions are first class objects which means that they can be passed as args to other functions. Functions can also be nested.
First class means that functions can be operated on like any other R object.
I.e., ...this means the language supports passing functions as arguments
to other functions, returning them as the values from other functions,
and assigning them to variables or storing them in data structures...
(Wikipedia article "First class functions").
Function arguments evaluated lazily, which means that they are only evaluated as they are needed/used.
Functions are declared as follows and return whatever the last statement evaluates to...
five <- function() {
+ 1 + 4
+ }
> five()
[1] 5
Like many scripting langauges, function parameters can be declared too, and with default parameters
> threshold <- function(x, n=5) {
+ x[x > n] = n
+ x
+ }
> threshold(1:10, 2)
[1] 1 2 2 2 2 2 2 2 2 2
Also like many other languages, functions can accept a variable number of arguments, using the C-like syntax of three consequitive dots, or "ellipsis". For more information read this r-bloggers article.
The '...' argument indicates variable number of arguments. You can pass the list using '...' to other functions as well. Note that arguments after '...' must be named explicitly and cannot be partially matched.
> test <- function(...) {
+ arguments <- list(...)
+ arguments
+ }
> test(1,2, a=3)
[[1]]
[1] 1
[[2]]
[1] 2
$a
[1] 3
If you want to get a lit of the formal arguments a function expects,
use the formals(functionName) function, which returns a
list of all formal argumentss of the function with name "functionName"
Scoping
Best read the R manual scope section.
To bind a value to a symbol, R searches though series of environments.
An environment is a collection of symbol, value pairs. To find out what is
in a function's environment use the ls(funcName) function.
Each environment has a parent and a number of children. A function plus
an environment is a closure.
From the command line R will...
- Search global environment (user's workspace) for the symbol name,
- Search the namespaces of each package on the search list (order matters).
You can print search list using the
search()function.
R has seperate namespaces for objects and functions so functions and objects can have the same name without a name-collision occuring.
Scoping rules determine how a value is associated with a
free variable in a function. R uses lexical scoping, also known as static scoping.
In lexical or static scoping the value associated with a variable is
determined by its lexical position (i.e, where in the text/code structure with
respect to nesting levels, functions etc the variable is), which means that
... this matching only requires analysis of the static program text
.
This is the opposite of dynamic scoping wherethe value is determined by
the run time context.
Another way of saying this is that in lexical scoping the value of a variable in a function is looked up in the environment in which the function was defined. With dynamic scoping, a variable is looked up in the environment from which the function was called (sometimes refered to as the calling environment).
Free variables are those that are not formal arguments and are not local variables defined inside the function body.
R will search for free vars in the current environment and then recursively up into
the environment's parents until global environment is reached.
Then R will search the search() list.
A free variable in a function can be read and written to and will refer to some variable outside of the function's scope. The trouble is that free variables automatically become (are considered as) local variables if they are assigned to.
So, consider this little rubbish example:
> b <- 10
> a <- function() {
+ print(b)
+ }
> a()
[1] 10
Here b is a free variable in the function a() because
it is not declared as a parameter and it is not local.
But wait a minute... if we assigned a value to b rather than
just read it, it would become a local variable. There would be two b's.
One in the function's environment and one in the global environment and the
one in the function's environment would be modified!
This is why the <<- operator is needed!...
> a <- function() {
+ b <<- 100000
+ }
> a()
> b
[1] 1e+05
The <<- operator can be used to assign a value to an object in an environment that is different from the current environment. This operator looks back in enclosing environments for an environment that contains the symbol. If the global or top-level environment is reached without finding the symbol then that variable is created and assigned there.
See this SO thread, I found it very useful.
Vectors, Sequences And Lists
Vectors can only contain elements of the same type. When mixing objects coercion occurs so all vector elements are of the same type.
Sequences are just vectors of numerics with monotonically increasing values.
Lists are vectors that can contain elements of different classes.
Indices start from 1. If you index beyond the limits of
a list or vector you will get NAs.
Creation
Some easy ways to create 'em...
Vectors
vector(type, value)
For example, "vector("numeric", 10)" creates a
vector of 10 numerics all initialised to zero.
as.vector(obj)
Coerces obj into a vector. This will remove the names
attribute.
c(item1, item2, item3, ...)
The default form creates a vector by concatenating all the
items passed into c() together. All arguments
are coerced to a common type.
rep(item, times = x)
This will create a vector consisting of the item repeated
x times. For example...
> rep(123, times=5) [1] 123 123 123 123 123
rep(list-of-items, times = x)
This will create a flattened list of list-of-items
repeated x times. For example...
> rep(c(1,2,3), times=5) [1] 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3
In other words it is as if we keep appending each list to a "mother" list like
this: [[1 2 3] ... [1 2 3]] and then flatten the list
so that it becomes [1 2 3 ... 1 2 3]. Note that this
flattening is recursive so that lists of lists are
entirely flattened. For example...
> rep(c(c(1,2), c(2,3)), times=5) [1] 1 2 2 3 1 2 2 3 1 2 2 3 1 2 2 3 1 2 2 3
rep(list-of-items, each = x)
Like the above except that instead of pasting many copies of
the lists together, we first recursively flatten the list and then
repeat the first value x times, then the second value
x, then the third and so on...
> rep(c(1,2,3), each=5) [1] 1 1 1 1 1 2 2 2 2 2 3 3 3 3 3 ## Or equivalently ## rep(1:3, each=5)
Sequences
x:y
This generates a sequence of numbers from x to y inclusive in increments of 1.
seq_len(len)
Generates a sequence from 1 to len. Useful in
for loops...
> seq_len(10)
[1] 1 2 3 4 5 6 7 8 9 10
> for (i in seq_len(10)) {
+ # Is a loop of 10 iterations, with i values as above
+ }
seq(x, y, by=[0-9]+)
This is a generalisation of the above. It generates a sequence of numbers from x to y inclusive with a step of [0-9]+. Note that the inclusivity of the final value depends on the step value. For example...
> seq(1,5,by=3) [1] 1 4
seq(x,y,length=[0-9]+)
As above but we don't care about the increment but want x to y inclusive such that length of the generated sequence is [0-9]+. Note that x and y are always included. For example...
> seq(1,5,length=9) [1] 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0
seq(along.with=obj) or seq_along(obj)
The above are equivalent. I think you'd normally see seq_along()
rather than the longer form. This function enumerates the
object passed in. For example...
> s <- rnorm(10) ## Generate a vector of 10 random nums > seq_along(s) [1] 1 2 3 4 5 6 7 8 9 10
Lists
Remember that lists are like vectors that can contain elements of different classes.
list()
Use to create a list or even a list of lists.
For example, "list(1, "string", TRUE)" creates
a list of 3 lists, where each list has one member.
Subsetting \ Indexing
Always useful to read is the
R Language Definition for Indexing. This
is especiall useful to understand the difference between the
[[]] and [] indexing operations.
A vector is subset using square brackets (like in most scripting
languages) and uses a very similar notation. Indices start from
1. If you index beyond the limits of a vector you will
get NAs.
Slices
One thing to note is that unlike in Python's NumPy, slices are not
views into an array. In the following example, taking a slice of
vector a and modifying it does effect the original a.
> x = vector("numeric", 5)
> x[1:4] = 33
> x
[1] 33 33 33 33 0
> a = x[1:4]
> a
[1] 33 33 33 33
> a[1] = 22
> a
[1] 22 33 33 33
> x
[1] 33 33 33 33 0
Boolean Indexing
eg. y<- x[!is.na(x)]
Fancy Indexing
Can also fancy-index vectors:
x[c(1,3,5)] ## gets first, third and fith element x[-c(2,10)] ## gives us all elements of x EXCEPT for the 2nd and 10 elements! # ^ # Note the minus sign here!
Named Indexing
You can assign names to each index of a vector (or list) and then index it using that name instead of position. For example:
> x = vector("numeric", 2)
> x
[1] 0 0
> ## By default vector has no names
> names(x)
NULL
> ## Note how the vector is now displayed in a tabular fashion
> names(x) = c("john", "jack")
> x
john jack
0 0
> ## You can get the first member using its name
> x["john"]
john
0
> ## Or can get the first member using its position
> x[1]
john
0
With lists things get a little more interesting. You can give the
elements of a list names, just as we did above for vectors, but when
we do we can use the $ operator to access named
members: list$member.name.
> q = list(1, 1.2, "james", TRUE)
> names(q) = c("a", "b", "c", "d")
> q$a ## Now we can refer to the member using its name
[1] 1
> q$b ## Now we can refer to the member using its name
[1] 1.2
Single Item Or List: [[ vs [, or double brackets vs single brackets...
For lists, use [[ to select any single element. [
returns a list of the selected elements:
> x <- list(1,c(2,3)) > x [[1]] ## This is list element 1 [1] 1 ## List element 1 is a numeric vector of size 1 [[2]] ## This is list element 2 [1] 2 3 ## List element 2 is a numeric vector of size 2 > > x[1] ## Returns a list containing the selected numerics [[1]] [1] 1 > class(x[1]) ## Single '[' returned a list [1] "list" > > x[[1]] ## Returns the actual element [1] 1 > class(x[[1]]) [1] "numeric"
The other significant difference is that double brackets only let you select a single item, whereas, as you have seen, the single bracket lets you select multiple items.
Note that with vectors there is only a very subtle difference using
the indexing operations [[]] and []. If we
use a single index then in both cases an atomic type will be returned.
However, using [[]] will strip the names.
> # Use the single braces [...]
> x["james"]
james
0
> class(x["james"])
[1] "numeric"
> x["james"] + 1 # NOTE: The names() have been preserved
james
1
>
> Use the double braces [[...]]
> class(x[["james"]])
[1] "numeric"
> x[["james"]] + 1 # NOTE: The names() have been DROPPED
[1] 1
Test For Membership
To see if a value is in a list or vector use %in% or match().
> v <- c('a','b','c','e')
## %in% returns a boolean
> 'b' %in% v
[1] TRUE
## match() returns the first location of 'b', in this case: 2
> match('b',v)
[1] 2
One advantage that %in% has over match() is that,
because it returns a list of booleans, it can be used with functions
such as all() or any(). For example...
> all(c('b', 'e') %in% v)
[1] TRUE
Other Common Operations
length(seq_obj)
dim() attribute.
identical()
paste(list, collapse=str)
Collapses the list into a single string in which the elements
are separated by the string specified by str. The
elements in the list will be coerced into a string using
as.string() if required. For example...
> paste(c(1,2,3,4,5), collapse = "") [1] "12345"
You can, in fact, paste multiple lists into a string, the first element from each list first, then the second and so on. For example...
>paste(1:3, c("X", "Y", "Z"), 4:7, collapse="")
[1] "1 X 42 Y 53 Z 61 X 7"
Matrices
Intro
Matrices are like 2D vectors. All elements must be of the same type.
They have an extra dimensions attribute called dim which
gives the matrix shape as a tuple (num rows, num cols).
For example...
> # Creates a matrix initialised with NA values. > m <- matrix(nrow = 2, ncol = 3) > attributes(m) $dim [1] 2 3
Use dim() to get the dimensions of a matrix. Vectors do not
have a dim attribute... only matricies and data frames.
Use length() for a vector.
Constructing
There are several ways to construct a matrix...
By default hey are constructed column-wise: Fill the first column, then second, and so on.
This can be useful when your data is presented as a flat vector where every group of n items is a list of observations for one single variable.
For example, lets say that we have a vector that records the test scores
for 2 students across 4 exams. The vector could be a
flattened version of [[80, 81, 82, 83], [50, 51, 52, 53]],
where each inner vector is the test scores for a student in 4 seperate
exams.
The flattened version we have would be
[80, 81, 82, 83, 50, 51, 52, 53]. The first
student has done very well, scoring in the 80's. The second student
has only scored in the 50s (just for the sake of making distinguishing
students easy). We want a matrix where columns represent a student
and each row the score for a particular test. I.e, we want
a matrix where the first colum had the values 80, 81, 82, 83
and the second column had the values 50, 51, 52, 53. To read
the vector into a matrix, column-wise, we would do the following.
> data = c(80, 81, 82, 83, 50, 51, 52, 53)
> data
[1] 80 81 82 83 50 51 52 53
> matrix(data, nrow=4, ncol=2)
[,1] [,2]
[1,] 80 50
[2,] 81 51
[3,] 82 52
[4,] 83 53
The above shows how we have given a shape to our flat vector of data. Now we have students in the matrix columns and tests as the rows.
It is also possible to fill the matrix row-wise: Fill the first row, then the second, and so on.
Imagine our flat vector from above was actually presented in a different
way. The consecutive values are now organised per test so that the
vector, unflattened, would look like this: [[80, 50], [81, 51],
[82, 52], [83, 53]]. We still want our matrix layout to be
a column for each student and the rows to be the tests. What we need
to do is fill by row, as follows:
> data = c(80, 50, 81, 51, 82, 52, 83, 53)
> m <- matrix(data, nrow=4, ncol=2, byrow=TRUE)
> m
[,1] [,2]
[1,] 80 50
[2,] 81 51
[3,] 82 52
[4,] 83 53
Another way to accomplish exactly the same thing as we have done above
is to simply apply the dim attribute to our vector. Note
however that doing it this we way are restricted to fill-by-column!
> data <- c(80, 81, 82, 83, 50, 51, 52, 53)
> data
[1] 80 81 82 83 50 51 52 53
> dim(data) = c(4, 2)
> data
[,1] [,2]
[1,] 80 50
[2,] 81 51
[3,] 82 52
[4,] 83 53
Matrix Subsetting
Use matrix[rowIdxs, colIdxs]. If either index ommited
(must still include the comma) then all of that index is returned.
The defualt for single matrix element is to return it as a vector. To
stop this use drop=FALSE. Same issue for subsetting a single
row or column.
x <- matrix... x[1,2] ##< returns vector x[1,2, ##< drop=FALSE] returns a 1x1 matrix
Getting Matrix Info
To get the shape of a matrix, as we have seen, we can use
dim() which returns a tuple (num rows, num cols).
To get just the number of rows use nrow().
To get just the number of columns use ncol.
Combining Matricies
Use cbind() or rbind().
Maths With Matricies
"Normal " Uses Broadcasting: It's Not Matrix Multiplication!
Much like most other vectorised languages like Yorick or Python's NumPy, R supports broadcasting. This means we can do things like multiply a matrix (or indeed a vector) by a scalar and the result is the multiplication applied element-wise:
> data <- c(80, 81, 82, 83, 50, 51, 52, 53)
> data * 10
[,1] [,2]
[1,] 800 500
[2,] 810 510
[3,] 820 520
[4,] 830 530
You can also multiply a matrix by a vector and the broadcasting will will work in the same element-wise fashion down the columns of the vector. The vector you are multiplying doesn't even have to have the same length as the column. As long as the column length is a multiple of the vectgor, R will continually "reuse" the vector:
# Here the vector has the same length as the matrix column.
> data * c(10,1,100,1000)
[,1] [,2]
[1,] 800 500
[2,] 81 51
[3,] 8200 5200
[4,] 83000 53000
# The vector is shorer than the column length, so R reuses the vector
# essentally multiplying by c(10, 1, 10, 1)...
> data * c(10,1)
[,1] [,2]
[1,] 800 500
[2,] 81 51
[3,] 820 520
[4,] 83 53
We can also multiply a matrix by another matrix. But beware, the standard multiply symbol, *, does NOT do matrix multiplication as we see here:
> m < rep(10, 8)
> dim(m) = c(4,2)
> m
[,1] [,2]
[1,] 10 10
[2,] 10 10
[3,] 10 10
[4,] 10 10
> data * m
[,1] [,2]
[1,] 800 500
[2,] 810 510
[3,] 820 520
[4,] 830 530
Propper Matrix Multiplication!
To do an actual matrix multiplication in R you must use the%*%
operator.
Factors
What are they?
Factors represent catagorical data. This is data that can only be one of a set of values. For, example, the sex of a test subject can only be male or female.
They're kinda like a C enum type where each category has an
associated integer value that is unique to that category's label.
A factor has "levels". Each level is just one of the
enum labels. So, for example, if we had a list of participant
genders and converted it to a factor variable we would see the following:
> x <- factor(c("male", "male", "female", "male", "female", "female"))
> x
[1] male male female male female female
Levels: female male
We see that x is a list of gender labels, but has only 2 "levels", namely "male" and "female".
We can count the number of occurences of each level using the table()
function as follows:
> table(x) x female male 3 3
You can set the order of levels in a factor as well. Sometimes this is useful when plotting. You can also reorder factors if you need to.
> x <- factor(c("male", "male", "female", "male", "female", "female"), levels=c("male", "female"))
> x
[1] male male female male female female
Levels: male female
## ^^
## Note, how compared to prev example, the order of the levels is changed
Creating Factor Variables
## Create from a list of strings
> x <- factor(c("male", "male", "female", "male", "female", "female"))
> x
[1] male male female male female female
Levels: female male
## Assign levels to lists
> factor(c(TRUE, FALSE, TRUE), labels=c("label1", "label2"))
[1] label2 label1 label2
Levels: label1 label2
## Generate Factor Levels Using gl(num-levels, num-reps, ...)
> gl(2, 5)
[1] 1 1 1 1 1 2 2 2 2 2
Levels: 1 2
> gl(2, 5, labels=c("jeh", "tech"))
[1] jeh jeh jeh jeh jeh tech tech tech tech tech
Levels: jeh tech
Ordered factors
Ordered factors are factors but with an inherent ordering or rank: used for ordinal data.
From the R help files:
Ordered factors differ from factors only in their class, but methods and the model-fitting functions treat the two classes quite differently
.
What it is saying is that these functions will operate on and present the
data in the order that is specified by the ordered factor.
For example, let's create a dummy little factor set:
> ordered(1:5)
[1] 1 2 3 4 5
Levels: 1 < 2 < 3 < 4 < 5 # NOTICE THE "<" characters:
# These show the ordering of the factor.
The same effect can be produced using factor(1:5, ordered=TRUE).
Interactions
The "interaction" of two or more factors is basically their
cartesian product. Use the interaction() function
to create these interations.
Missing Values
Sadly most data sets contain a lot of missing data. In R missing values
are generally represented by NA or NaN. Whilst both
can represent missing data there are some subtle differences:
NA is a constant which is a missing value indicator. NA values
can have a class (i.e., be integer, char etc). NA is not the
same as NaN.
> is.na(NA) [1] TRUE > is.nan(NA) [1] FALSE
NA
Note that comparing against NA, e.g, vector_var == NA will
just give you a vector of NAs not booleans! This is because NA is not
really a value, but just a placeholder for a quantity that is not available.
This also means that something like x[!is.na(x) & x > 0] != x[x > 0]
because if x contains NAs x[x > 0] will return a list containing
the NAs as well as those values greater than zero. This is because NA
is not a value, but rather a placeholder for an unknown
quantity and the expression NA > 0 evaluates to NA.
NaN
NaN means "Not a Number" NaN are also NA but do not
have a class:
> is.na(NA) > is.na(NaN) [1] TRUE > is.nan(NaN) [1] TRUE
Tabular Data: Data Frames
The trouble with a matrix is that all the data must be of the same type and most of the time one will be dealing with tables of results where the columns have very different types.
Data frames to the rescue. The are like tables where each column
can have its own type. On a technical level, a data frame is a list, with the components of
that list being equal-length vectors
[2].
Create A Data Frame
A Dataframe can be created manually using the data.frame
constructor although normally they are created by reading in data from
a data source such as a file (for instance, read.table() or
read.csv()). By default strings are converted to factors. This
can behaviour can be changed using stringsAsFactors=FALSE.
Manual Creation
To create a dataframe manually use the data.frame constructor.
Pass in many lists, all of the same length but possibly different types,
each of which will become a column in the table with the given name.
The table columns will be created in the order specified in the constructor
arguments...
> x <- data.frame(jeh = c("j", "e", "h"), tech = c(1,2,3))
> x
jeh tech
1 j 1
2 e 2
3 h 3
One thing that is interesting to note is that the values in column
"jeh" have been cooerced into factors, which is
apparent if you use str(x) as shown in the next section. To
override this default behaviour you can set stringsAsFactors = FALSE
int he constructor.
As mentioned, a data fram is a list of lists-of-equal-length. You can
revert from a dataframe back to a list of lists using as.list(df).
It will convert the datafame to a list of named vectors...
> as.list(x) $jeh [1] j e h Levels: e h j $tech [1] 1 2 3
Reading In Tabular Data
One thing to note: For large datasets estimate amount of RAM needed to store dataset before reading it in. The reason being that R will read your entire dataset into RAM. It does not support (at least without special packages) dataframes that can reside on disk.
Use object.size(df) to get the size of a dataframe in
memory. For pretty-print the results use
print(object.size(df), units="Mb").
read.table(file, header, sep, colClasses, nrows, comment.char, skip, stringsAsFactors)
## header - is the first line a header or just start of data
## sep - column delimeter
## colClasses - vector of classes to say what class of each col is
Not required but specifying this can increase speed
of operation.
data <- read.table("filename.txt")
## will auto figure out column classes, sep etc etc, auto skips
## lines with comment symbol etc etc.
read.csv is identical to read.table except default seperator is comma (,)
Information Above Your Data Frame
ncol(df) gives number of columns.
nrow(df) gives number of rows.
names(df) gives names of each column.
colnames(df) to retrieve or set column names.
rownames(df) to retrieve or set row names.
head(df, n=...)
tail(df, n=...)
summary(df) gives some info for every variable. For example,
looking at the dummy table created above gives:
> summary(x)
jeh tech
e:1 Min. :1.0
h:1 1st Qu.:1.5
j:1 Median :2.0
Mean :2.0
3rd Qu.:2.5
Max. :3.0
str(df) compactly displays the internal structure of an R object.
> str(x) 'data.frame': 3 obs. of 2 variables: $ jeh : Factor w/ 3 levels "e","h","j": 3 1 2 $ tech: num 1 2 3
quantile(df$col, na.rm=T/F) looks at quantiles. Can pass
probs=c(1,2,3) for different percentiles.
table(df$col) to make a table summarising the column by grouping
occurences. Can pass two variables to make it 2D table.
as.list(df) will convert the datafame to a list of named
vectors.
To get a list of column types use the following:
> coltypes <- as.character(lapply(x, class)) > coltypes [1] "factor" "numeric"
... or equivalently ...
coltypes <- sapply(x, class)
Indexing Your Data Frame
As previosly said, ... a data frame is a list, with the components of
that list being equal-length vectors
[2]. I.e., each
column is a list. Therefore the
list-accessing operators will work
here:
Subsetting Columns
> x <- data.frame(jeh = c("j", "e", "h"), tech = c(1,2,3))
> ## Get a single column using numeric index
> ## Because x is a list of lists the [[...]] operator returns a list
> x[[1]]
[1] j e h
> ## Get a single column using its name
> ## This will also return a list
> x$jeh
[1] j e h
> ## Get a single column as a dataframe
> ## Because the single '[' returns a list of the contents it essentially
> ## returns a list of lists which R is clever enough to know as a data
> ## frame
> x[1]
jeh
1 j
2 e
3 h
> ## Because x is a list we can use all list indexing ops like
> ## fancy indexing
> x[c(1,2)]
jeh tech
1 j 1
2 e 2
3 h 3
> ## Or boolean indexing
> x[c(FALSE, TRUE)]
tech
1 1
2 2
3 3
> ## Or use ranges
> x[1:2]
jeh tech
1 j 1
2 e 2
3 h 3
Note that in the above examples for fancy and boolean indexing we had
to enclose the indexers with c(...). This has to be done,
otherwise R will think you are trying to index multi-dimensions!
Subsetting Rows
To subset the rows of a dataframe use a 2D index of the form
[row, col]. Each index specifier can be any of the
list indexers like ranges, fancy indices, boolean arrays etc.
For example, x[condition1 & condition2,]
(note the trailing comma) would only select certain rows and all
columns.
To get indices of selected elements rather than boolean arrays,
use which(). It also auto removes all NAs.
Subset returns a list or dataframe...
When the subset returns a portion of one single column the result returned is generally a list:
> x[1:2, 1] [1] j e Levels: e h j
It might be your want it returned as a dataframe. If so specify
drop=FALSE as a kind-of 3rd dimension...
> x[1:2, 1, drop=FALSE] jeh 1 j 2 e
Check for and filter out missing values
You can find missing values using the following methods...
> ### Using is.na() > sum(is.na(df$col)) ## == 0 when no NAs > any(is.na(df$col)) ## == FALSE when no NAs > all(colSums(is.na(df)) == 0) ## == FALSE when no NAs for entire df
You can also use is.na() to filter out NAs in a list or dataframe
column:
> bad <- is.na(a_list) a_list[!bad]
But, to filter out all dataframe rows where any column has an NA value, use the following...
> ## Rows where NO column has an NA - complete.cases() > y <- rbind(x, c(NA, 4)) > y jeh tech 1 j 1 2 e 2 3 h 3 4 NA 4 ## << Note the NA here > good <- complete.cases(y) y[good,] jeh tech 1 j 1 2 e 2 3 h 3
Splitting a data frame
The function split() divides the data in a list or vector into groups identified by a factor. Given that a Data Frame is just a list of lists, we can use split() to divide up the table into many sub-tables (like "groups"). Often this is the first stage in a split-apply-combine problem.
A really noddy example...
f = factor(rep(c("A", "B", "C"), each=3))
> f
[1] A A A B B B C C C
Levels: A B C
> x = data.frame(a = 1:9, b = f)
> x
a b
1 1 A
2 2 A
3 3 A
4 4 B
5 5 B
6 6 B
7 7 C
8 8 C
9 9 C
> split(x, x$c) $A a b 1 1 A 2 2 A 3 3 A $B a b 4 4 B 5 5 B 6 6 B $C a b 7 7 C 8 8 C 9 9 C
The rather crummy example above shows how split() has essentially divided up the data frame into groups defined by the column 'b'.
To reduce typing a little and increase the readability, when the data frame and column names are long you can use with() to replace "split(df, df$col)" with "with(df, split(col))".
Be careful when thinking of split() as doing a group-by, however, because it is not doing this... it is doing a more naive split of the data as we can observe if we modify the call to split() in the above example:
> split(x, factor(c("X", "Y", "Z")))
$X
a b
1 1 A
4 4 B
7 7 C
$Y
a b
2 2 A
5 5 B
8 8 C
$Z
a b
3 3 A
6 6 B
9 9 C
We can see this visually in the following diagram which demonstrates how split is working and also how it will recycle the factor by which it is using to split on...
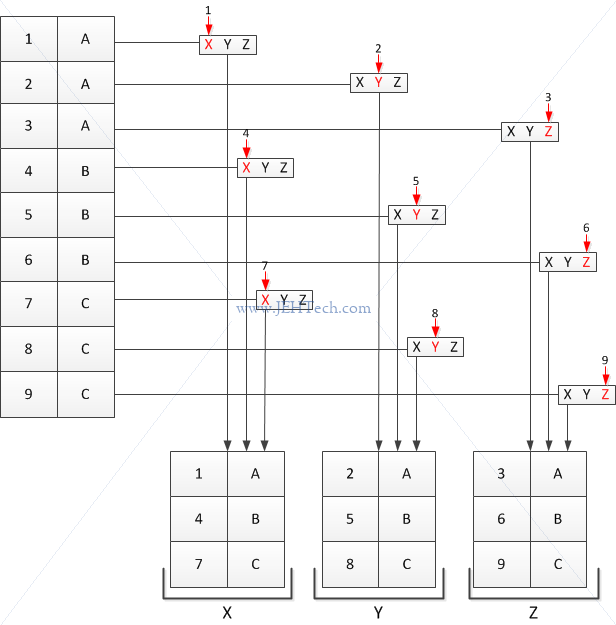
So, as you can see, it is not really a grouping operating. It is really just flinging each ith row into a bucket with the name of the corresponding ith factor variable, and if there are not enough factor variables then they are recycled. That is why it is only acting like a group-by when we pass in a data frame and a row of the data frame as the factor to split on.
Okay, great, but what's the use in this. Well usually we would, instead of splitting a data frame in its entirity, we would split a column we are interesting in calculating some statistics over based on a grouping re another column. In the above example, we might want to find the mean of the column "a" for each of the groups "A", "B" and "C". We would use the following:
> with(x, lapply(split(a, c), mean)) $A [1] 2 $B [1] 5 $C [1] 8
The rest of the notes
Find some training data at http://archive.ics.uci.edu/ml/datasets.html. http://data.gov.uk/data/search. DATA FRAMES ----------- x[row, col] with indices or actual names or slices or boolean conditions eg x[condition1 & condition2,] -- select only certain rows which() returns indices and doesnt return the NAs sort(x$colname, decreasing=TRUE/FALSE, na.last=TRUE) x[order(x$colname1, x$colname2, ...),] - order rows so column(s) in order plyr package for ordering arrange(dataframe, colname) -asc order arrange(dataframe, desc(colname)) - same with decreasing order add row/col ----------- df$new_col <- ... vector or use cbind() command - column bind or library(plyr) mutate(df, newvar=...) - adds new col and returns new datafame copy. TODO: Cross tabs (xtabs) TEXTUAL FORMATS --------------- dump() and dput() saves metadata like column class. still textual so readable but meta data included textual data works better in version control software but not space efficient y <-data.frame(...) dput(y) prints to screen dput(y, "filename.R") saves to file y2 <- dget("filename.R") loads DF back into Y2 Looks like little bit like object pickling in Python dget() can only be used on a SINGLE R object. dump() can be used on multiple objects, which can then be read back using a single source() command eg. x <- ... y <- ... dump(c("x", "y"), file="...") // pass NAMES of objects in rm(x, y) source("...") // restores x and y CONNECTIONS TO THE OUTSIDE WORLD -------------------------------- file gzfile (gzip) bzfile (bzip2) url (webpage) above function open connectsion to a file or a web page con <-file("foo.txt", "r") data <- read.csv(con) close(con) same as data <- read.csv("foo.txt") con <- url("http://....") x <- readLines(con) // x will be a character vector holding page source DATES & TIMES -------------- Dates - Date class Stored internally as days since 1-1-1970 Times - POSIXct or POSIXlt class Stored internallay as seconds since 1-1-1970 POSIXct just a large integer POSIXlt is a list storing day, week, day of year, month, etc etc Convert strings to dates using as.Date() x <- as.Date("1970-01-02") unclass(x) == 1 Generic functions: weekdays() - Give day of the week months() - Gives month name quarters() - Gives "Q1", "Q2" ... x <- Sys.time() p <- as.POSIXlt(x) p$sec == ..seconds.. strptime() converts dates to an object eg datestring <- c("January 10, 2013, 10:30") x <- strptime(datestring, "%B %d, %Y %H:%M") x will be a POSIXlt object ?strptime for help on function Normal +,-,<,> etc work on dates The operators keep track of leap years, leap seconds, daylight savings and timezones for us automatically :) LOOP FUNCTIONS -------------- All implement the Split-Apply-Combine strategy: SPLIT into smaller pieces, APPLY a function to each piece, COMBINE the results. lapply() - loop over list and eval func for each element. looping done in C split() - splits objects into sub-pieces lappy(list, function_name, ) returns result for every object in the list as a list in the same order. 'function_name' is applies to each element of the list and the return value used for that element in the new list. The arguments will be passed to your function_name(). lappply() generally uses ANONYMOUS FUNCTIONS. lapply(x, function(params){...}) sapply() - simplifies result of lapply(). Each list might go to vector if every element has length one. If cant figure out the simplification a list is returned. apply(X, margin, fun,...) margin is an int vector indicating which margins should be retained fun is func to be applied passed to 'fun' the margin refers to the DIMENSION number being used. For a matrix 1 is rows 2 is columns. So. apply(x,2,mean) would apply mean() to columns of matrix. For row/col sums/means use more highly optimized functions for SPEED rowSums = apply(x,1,sum) colSums = apply(x,2,sum) can keep two dims using appy(x, c(1,2,...), ) for multi dim arrays or use rowMeans(..., dim=x) mapply(fun, , MoreArge=NULL, SIMPLIFY=TRUE, USE.NAMES=TRUE) Applies a func in parallel over a set of different arguments. Eg two lists where element x in list_1 is param of 'fun' and element x in list_2 is second param. this is what mapply helps you do. Use to VECTORIZE FUNCTIONS that normally act on scalars. tapply(x, index, fun, ...) - apply a function over subsets of a vector. index is a factor or a list of factors which identifies which group each element of the numeric vector is in. e.g. x <- c(rnorm(10), runif(10), rnorm(10,1)) f <- gl(3,10) // creates a factor variable, each one repeated 10 times == 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 3 3 3 3 3 3 3 3 3 3 levels: 1 2 3 tapply(x, f, mean) 1 2 3 0.114 0.516 1.246 if you dont simply the result you get back a list You can subset a matrix to do the equivalent of an SQL GROUP BY: with(df, tapply(col1, col2, mean)) This will group by col1 and take the mean of col2 using that grouping. split(x, f, drop = false, ) like tapply() it splits x into groups where f identified which group each element of x is in. common to split() something then apply a function over the groups: lapply(splot(x,f), mean) for example split() can be used on complex data structures like dataframes. e.g. s <- split(dataframe, dataframe$col_name) lapply(s, function(x) snip ) tip: use sapply(... na.rm=TRUE) To split on more than one level, combine factors using interaction(): x <- rnorm(10) f1 <-gl(2,5) # 1 1 1 1 1 2 2 2 2 2, levels 1 2 f2 <-gl(5,2) # 1 1 2 2 3 3 4 4 5 5, levels 1 2 3 4 5 interaction(f1,f2) #10 levels 1.1 2.1 1.2 2.2 1.3 2.3 1.4 .. 2.5 > f1 <-gl(2,5) # 1 1 1 1 1 2 2 2 2 2, levels 1 2 > f1 [1] 1 1 1 1 1 2 2 2 2 2 Levels: 1 2 > f2 <-gl(5,2) > f2 [1] 1 1 2 2 3 3 4 4 5 5 Levels: 1 2 3 4 5 > interaction(f1,f2) 1 1 1 1 1 2 2 2 2 2 . . . . . . . . . . 1 1 2 2 3 3 4 4 5 5 | | \/ [1] 1.1 1.1 1.2 1.2 1.3 2.3 2.4 2.4 2.5 2.5 Levels: 1.1 2.1 1.2 2.2 1.3 2.3 1.4 2.4 1.5 2.5 NOTE: Although it says there are all those levels it appears it uses the returned array, not the levels themselves which is why it returns missing values for some levels so it has all the levels but not all are used. split(x, list(f1,f2), drop=TRUE) $'1.1' [1] 2.0781323 0.7863726 $'1.2' [1] -0.7285167 0.1410324 $'1.3' [1] 0.643741 $'2.3' [1] -0.7616546 $'2.4' [1] -0.4612373 -0.6063274 $'2.5' [1] 0.0106792 -0.5811911 split(drop = TRUE) to drop empty levels Some useful functions ---------------------- range() returns the minimum an maximum of its argument unqiue() returns vector with all duplicates removed grep, grepl, regexpr, gregexpr and regexec search for matches to argument pattern within each element of a character vector DEBUGGING --------- Return something invisibly: return the value but dont print it use invisible(x). 5 basic functions traceback() prints function call stack debug() flags function for debug mode which lets you step through eg debug(func) func() Then drops into browser "n" to step through to next instruction browser() suspends execution wherever it is called and puts func into debug mode trace() allos insert debugging code into function at specific places recover() allows modification of the error behabiour so you can browse the function call stack options(error = recover) R Profiling ----------- Figure out why things are taking time and suggest strategies for fixing. Systematic way to examine how much time is being spent in various parts of your program. system.time() ------------- Takes R expression (can be in curley braces) an returns time taken to do the expression in seconds. Returns proc_time user time - CPU time elapsed time - "wall clock" time There are multi-threaded BLAS libaries (ATLAS, ACML, MKL) Parallel package for parallel processing http://stackoverflow.com/questions/5688949/what-are-user-and-system-times-measuringThe "user time" is the CPU time charged for the execution of user instructions of the calling process. The "system time" is the CPU time charged for execution by the system on behalf of the calling process.Rprof() -------- R must be compiled with profiler support. Rprof() starts the profiler DO NOT USE system.time() with Rprof() summaryRprof() to summarise Rprof() output by.total - divide time spent in each function by total time by.self - divide time spend in each function minus time spent in functions above by total time. Rprof() tacks FUNCTION CALL STACK at regularly sampled intervals Generate (pseudo) random numbers --------------------------------- - rnorn() random normal with mean and stddev - dnorm() evaluates standard normal prob density funct at point or vector of points - pnorm() eval cumulative distribution function for normal distribution - rpois() generate random Poisson with a given rate - rbinom() uses binomial distribution d for density r for random number gen p for cummulative distribution q for quantile function Important: use set.seed(<seed>) this resets the sequence that will occur. Random sampling ---------------- THe sample() function draws randomly from a specified set of (scalar) objects allowing you to sample from arbitrary distributions. set.seed(...) sample(1:10, 4) # without sample replacement sample(1:10, 4, replace=TRUE) # with sample replacement # (so can get repeats) READ DATA FROM WEB (HTTPS) ------------------------- library(RCurl) url <- "https://d396qusza40orc.cloudfront.net/getdata%2Fdata%2Fss06hid.csv" data <- getURL(url, ssl.verifypeer=0L, followlocation=1L) writeLines(data,'temp.csv') get binary data using getBinaryURL(...) and the following: tf <- tempfile() writeBin(con=tf, content) READ DATA FROM EXCEL -------------------- library("xlsx") dat <- read.xlsx(fname, startRow=18, endRow=23, colIndex=7:15) XML into R --------------- library(XML) doc <- xmlTreeParse(url, useInternal=TRUE) there is also htmlTreeParse for HTML files! rootNode <- xmlRoot(doc) xmlName(rootNode) xmlName() - element name (w/w.o. namespace prefix) xmlNamespace() xmlAttrs() - all attributes xmlGetAttr() - particular value xmlValue() - get text content. xmlChildren(), node[[ i ]], node [[ "el-name" ]] xmlSApply() xmlNamespaceDefinitions() xmlSApply(node, function-to-apply) - recursive by default or xpathSApply(node, xpath-expression, function-to-apply) XPath language /node - top-level node //node - node at any level node[@attr-name] - node that has an attribute named "attr-name" node[@attr-name='xxx'] - node that has attribute named attr-name with value 'xxx' node/@x - value of attribute x in node with such attr. FAST TABLES IN R ----------------- data.table Is a child of data.frame but is much FASTER. Has a different syntax Create in exactly same way is data.frame tables() - all data.tables in memory Subsetting with only one index subsets with ROWS Subsetting columns very different. EXPRESSIONS Uses expressions: satements between curley brackets {} e.g. dt[, new_col := {some expressions...} LISTS Or use a list of functions to apply to columns: e.g. dt[, list(mean(x), sum(z))] ADD NEW COL (very efficiently) with := syntax dt[,w:=col-name] Adds inplace Data tables copied BY REFERENCE so be careful of aliases! Create copied explicitly using copy() function Grouping using 'by=' syntax. Special variable .N to count group eg. dt[, .N, by=...] Keys for fast indexing. joins, merges, subsetting, grouping e.g. setkey(dt, col-name) Apparently even faster is http://www.inside-r.org/packages/cran/data.table/docs/fread MYSQL ----- dbCon <- dbConnect(MYSQL(), user="...", host="...", db="...") allTabls <- dbListTables(dbCon) result <- dbGetQuery(dbCon, "SQL QUERY STRING") dbListFields(dbCon, "TABLE NAME") - lists field names in table dbReadTable(dbCon, "TABLE NAME") - reads the ENTIRE table (caution - size!!) To select a subset: query <- dbSendQuery(dbCon, "SQL STR") - doesn't get the data x <- fetch(query, n=number-of-rows) - this gets the data dbClearResult(query) - must be done after a query! dbDisconnect(dbCon) - must be closed!!!! see on.exit() function for safety http://www.r-bloggers.com/mysql-and-r/ HDF5 ---- source("http://bioconductor.org/bioLite.R") biocLite("rhdf5") library(hdf5) f = h5createFile("...") g = h5createGroup("file-name", "group-name") h5ls("file-name") - dump out file info Write data: A = matrix(1:10, nr=5, nc=2) h5write(A, "filename", "group/path/to/var/spec") will automatically write meta data for you too attr(A, "somejunk") <- "a value" See bioconductor notes FROM WEB -------- con = url("...") htmlCode = readLines(con) close(con) htmlCode is just the raw code as one blob. Alternative use htmlTreeParse() and xpathSApply() etc creating new variables ======================= create sequence ---------------- s1 <- seq(min-val, max-val, by=incr-value) s1 <- seq(min-val, max-val, length=) - has length values s1 <- seq(along = x) vector of indices for x create binary vars -------------------- x <- ifelse(df$col <op> cond, TRUE, FALSE) create categories ----------------- apply to quantative variables x <- cut(df$col, breaks=....) The breaks specifies the boundaries for the groups eg cut(df$col1, breaks=quantile(df$col2)) or use library(Hmisc) cut2(df$col, g=4) - shortcut for quantiles reshape data ============== USE THE reshape2 library library(reshape2) tidy data 1. every variable its own col 2. every observation its own row 3. each table/file to store data about only one kind of observation melt a dataframe ----------------- melt(df, id=c(...), measure.vars=c(...)) create a new df. variables in id list are repeated for each variable in measure.vars so that rather than being wide the df becomes thin with a row for each variable with ids acting like the index or identifier. Thus is there are 2 items in measure.vars there will be 2 occurances of the identifiers in id list. cast data frame ---------------- using dcast() to summarise a dataset - - - - - - - - - - - - - - - - - - - dcast(df, col-to-group-by ~ col-to-summarise) will take col-to-group-by and produce a row for each unique value in that set. then it groups by those values and applies, by default, a count to see how many instance of col-to-summarise belong to that group. if you dont want to count, you can apply any aggregation operation dcast(df, col-to-group-by ~ col-to-summarise, mean) split-apply-combine problems ------------------------------ Notes on this article: www.r-bloggers.com/a-quick-primer-on-split-apply-combine-problems Further note by Hadley Wickham list_by_group <- with(df, split(col-to-count, col-to-group-by)) aggr_by_group <- sapply(list_by_group, aggregation-function) e.g. aggregation-function could be mean() This can be summarised in one command in these different ways: 1. with(df, tapply(col-to-count, col-to-group, agrregate-function)) The tapply() function does an inherent group by operation for you 2. with(df, by(col-to-count, col-to-group, agrregate-function)) 3. aggregate(col-to-count ~ col-to-group, df, agrregate-function) dplyr package =============== library(dplyr) Basic verbs used by package: select - returns a subset of columns of df filter - extract subset of rows arrange - reorder rows rename - rename a column mutate - add new variables or columns. transform existing group_by - pipeline - use %>% to pipe output to next func. then funcs won't need the df argument, they get it from the pipeline join - faster but less fully featured than merge(). this can only merge on common names, you can spec the cols! join_all - join multple df's. Takes list as argument summarise - Format generally: new_df<- dplyr-func(df, what-to-do-with-df) select ------ Can use column names in ranges rather than just indices select(df, col-name1:col-name2) - select all columns in this range select(df, (-col-name1:col-name2)) - select all columns EXCEPT those in this range filter ------- filter(df, col <op> cond) arrange ------- df <- arrange(df, col-name) - order dataset by this column, asc order df <- arrange(df, desc(col-name)) - order dataset by this column, desc order rename ------ df <- rename(df, col-name-new=col-name-old, ...) - renames those cols, leaves others as is mutate ------ transform existing vars or create new ones df <- mutate(df, new.col=....) - adds a new column eg df <- mutate(df, new.col=old.col - mean(old.col)) - adds a new column group_by --------- grouped_df <- group_by(df, colname) summarise(grouped_df, label1=func(col-name), ....) merging data ============= By default merges by columns with common name. Use by.x, by.y to tell it which cols by.x is the first df by.y is the secon df use "all=TRUE" for an outer join merge(df_x, df_y, by.x=, by.y=, all=) can also use join in plyr package Misc ---- remove the original data frame from your workspace with rm("variable-name") packageVersion("package name") to get the package version Manipulating Data with dplyr ---------------------------- FIRST, you must load the data into a 'data frame table' data_df_tbl < tbl_df(data_df) dplyr supplies five 'verbs' that cover most fundamental data manipulation tasks: select(), filter(), arrange(), mutate(), and summarize(). dplyr also doesn't print all teh data. to see it all use View() (best done in R Studio) select(data_df_tbl, colname1, colname2,...) works on COLUMNS returns a new df_tbl with only the cols named in the order specified given this ordering, select can then allow us to use the range operator ":" with column names rather than indices. e.g. select(data_df_tbl, colname4:colname6) the order can be reversed as in colname6:colname4 too. to specify column to dump rather than keep use a minus "-". eg. select(data_df_tbl, -colname1) gives all columns except colname1 eg. select(data_df_tbl, -(colname1:cn3)) gives all columns except those in the range specified note the range must be enclosed in brackets to negate the entire range (rather than just the start element) Special select functions (from helpfile): - starts_with(x, ignore.case = TRUE): names starts with x - ends_with(x, ignore.case = TRUE): names ends in x - contains(x, ignore.case = TRUE): selects all variables whose name contains x - matches(x, ignore.case = TRUE): selects all variables whose name matches the regular expression x - num_range("x", 1:5, width = 2): selects all variables (numerically) from x01 to x05. - one_of("x", "y", "z"): selects variables provided in a character vector. - everything(): selects all variables. filter(data_df_tbl, boolean-vector-selecting-rows, boolean-vector... ) works on ROWS (the bool vectors are ANDed. To OR use "|" not "," ) eg. to select rows where a column value is not NA use: filter(data_df_tbl, !is.na(col-name)) arrange() orders the ROWS of a dataset according to a particular variable. eg to order ascending arrange(data_df_tbl, colname) eg to order descending arrange(data_df_tbl, desc(colname)) Can arrange based on multiple variables, sort my left most first, then next continuing right. mutate() Creates a new variable based on the value of one or more variables already in a dataset. eg. mutate(data_df_tbl, col_new = someFunc(col_existing)) Can also use values created in col_new's to the left, in col_new's to the right eg. mutate(data_df_tbl, col_new = someFunc(col_existing), col_new2 = col_new * 0.25) summarize() collapses the dataset to a single row, especially when data is grouped eg summarise(data_df_tbl, some-summary=func(col-name)) most useful with the group_by() verb group_by() When using summarize() on a table that has been group_by()'ed it will return the summary functions for EACH group rather than just one value :) merge() By default merges by columns with common name. Use by.x, by.y to tell it which cols by.x is the first df by.y is the secon df use "all=TRUE" for an outer join merge(df_x, df_y, by.x="col1", by.y="col2", all=) ^ Note col names are in quotes otherwise you get Error in fix.by(by.x, x) : object 'col1' not found Some special functions: - n() The number of observations in the current group. (ie. num rows) This function is implemented special for each data source and can only be used from within summarise, mutate and filter. - n_distinct() Efficiently count the number of unique values in a vector Faster and more concise equivalent of length(unique(x)) quantiles ---------- | We need to know the value of 'count' that splits the data into the top 1% and bottom 99% of packages based on total downloads. In statistics, this is called the | 0.99, or 99%, sample quantile. Use quantile(pack_sum$count, probs = 0.99) to determine this number. > quantile(pack_sum$count, probs = 0.99) tidying data with tydr ----------------------- library(tidyr) http://vita.had.co.nz/papers/tidy-data.pdf gather() Use gather() when you notice that you have columns that are not variables Looking at the example from the help file > # From http://stackoverflow.com/questions/1181060 > stocks < data.frame( + time = as.Date('2009-01-01') + 0:9, + X = rnorm(10, 0, 1), + Y = rnorm(10, 0, 2), + Z = rnorm(10, 0, 4) + ) > head(stocks) time X Y Z 1 2009-01-01 0.3269115 0.7445885 6.257515 2 2009-01-02 -1.5729436 -3.3722382 2.050222 3 2009-01-03 -0.5257739 -1.2664309 -5.498621 4 2009-01-04 -1.4181633 -0.2352554 2.338179 5 2009-01-05 -0.6081603 -2.1707088 2.099759 6 2009-01-06 0.8347829 2.5455941 9.737100 > x = gather(stocks, stock, price, -time) > head(x) time stock price 1 2009-01-01 X 0.3269115 2 2009-01-02 X -1.5729436 3 2009-01-03 X -0.5257739 4 2009-01-04 X -1.4181633 5 2009-01-05 X -0.6081603 6 2009-01-06 X 0.8347829 What have we done here? In our original data set we had 3 columns. Each represented the price of a stock. We want to have one column for the stock type and one column for the price of that stock. Why do we want this? This is one of the principles of "tidy data". (see http://vita.had.co.nz/papers/tidy-data.pdf). Really what we are discussing is type-of-stock and price-of-stock at a time. Therefore there are 3 variables and each column should be a variable. As it stands the dataset columns represent the type-of-stock variable split over many columns. This isn't always a bad thing, as the article discusses, but it can be tidied to just have the variables as columns: time, stock and price. This is called "melting" the dataset. So... gather(stocks, stock, price, -time) The data argument, stocks, is the name of the original dataset. The key/value arguments, stock and price, give the column names for our new tidy dataset. Stock is the key, because it specifies all the column names, X, Y, and Z are to be collected under this new variable... they become like keys in a DB table. The value argument is the values of these columns that will be matched to the key. -time excludes time from the melt, because it is already a variable separate() separate one column into multiple columns spread() Spread a key-value pair across multiple columns. extract_numeric() Easy way to just get the numbers out of a string. eg if you have a numerical identified with a common textual prefix. STANDARD FILE DOWNLOAD STUFF ---------------------------- if(!file.exists(your-dir-name)) { dir.create(your-dir-name) } url < "https://....." download.file(url, destfile=your-file-name, method="curl") data < read.csv(your-file-name) On windows, however, I seem to need to do this library(Rcurl) if(!file.exists(your-dir-name)) { dir.create(your-dir-name) } url < "https://....." writeLines(getURL(url, ssl.verifypeer=0L, followlocation=1L), your-file-name) data < read.csv(your-file-name) EDITING TEXT VARIABLES ---------------------- tolower() - returns all lowercase toupper() - returns all uppercase strsplit(string, seperator). Remember to escape a character you need double backslash. eg. "\\." sub(str-to-search-for, replacement-str, string) - replace first instance only gsub() - replace all instances grep(regexp, str, value=T/F) - returns indices grepl() - returns boolean mask vector library(stringr) nchar(str) - number of chacacters substr(str, start-index, end-index). Indices from 1 and are inclusive paste(str1, str2, sep=" ") str_trim() - trim off spaces at start or end. Names of variables * All lower case when possible * Descriptive as possible * Not duplicated * Not have underscores, dots or whitespace Working with dates ------------------- date() - returns string od date and time Sys.Date() - returns a Date object. Can reformat using format() Date objects can be subtracted, added etc format(Sys.Date(), "format-string") where format-string is like a prinf expression with these special placeholders %d = day as number (0-31) %a = abbreviated week day name %A = full week day name %m = month number (two digits) %b = abbreviated month %B = unabbreviated month %y = 2 digit year %Y = 4 digit year as.Date(char-vector, format-string) turns character vectors into dates weekdays(date-obj) returns the week day as a string months(date-obj) returns the logn month name as a string julian(date-obj) returns number of days since epoch (1st Jan 1970) library(lubridate) Easy date manipulation Instead of as.Date(..., "format") you can use... ymd(...) which will search for year, month, day in *all* feasible formats mdy(...) will search for month, day, year in *all* feasible formats ymd_hms(...) date and time and so on... The functions also accept the timezone using the tz argument to the functions. wday(..., label=TRUE) == weekdays() in lubridate data resources --------------- http://data.un.org - united nations http://www.ons.gov.uk/ons/index.html- uk office for national statistics https://open-data.europa.eu - european data on EU http://www.data.gov - usa gov http://www.data.gov/opendatasites <--- look here http://www.data.gov.uk - uk data site http://www.gapminder.org http://www.asdfree.com - help on usa surverys http://www.infochimps.com/marketplace http://www.kaggle.com http://data.worldbank.org Plotting in R ==================================================================== boxplot(df$col, col="blue", breaks=100) breaks is the number of bars (or buckets) hist(df$col, col="green") rug(df$col) - puts a "rug" under the histogram abline(h=num) - draw a horizontal line abline(v=num, col="red", lwd=2) - draw a vertical line lwd is line width barplot() base plotting system - the oldest start with blank canvas and add things to it one by one plot(x,y) or hist(x) launches the graphics device plot() is generic... it can behave diff depending on the data passed to it. ?par for base graphipcs system parameters eg with(df, plot(col1, col2)) key plotting parms pch - the polotting symbol a number representing a symbol types. try example(points) on the command line lty - the line type lwd - line width as integer multuple col - colour xlab - the label for x axis ylab - the label for y axis legend par() used for GLOBAL graphics parameters: las - orientation of axis labels on plot bg - background coloour mar - margine size margine 1 is the bottom, then rotate CW for 2, 3, 4 oma - outer margin size mfrow - # plots per row, col (filled row wise) mfcol - # plots per row, col (filled col wise) base plotting functions plot() - scatter plot lines() - adds lines to a plot points() adds points to a plot text() add text labels to plot at specific x,y corrds title - add annotations to x, y axis labels, title, subtitle etc mtext - add arbirary text to margins axis - add axis ticks/labels multiple base plots: par(mfrow = c(rows, vols)) with(your-dataframe, { plot(some data in first plot) plot(plot some data in second plot) }) To setup plot without plotting anothing plot(x,y type="n") ^^^^ make a plot except for the data Then you can plot the data sets one by one. Launch screen device quartz() on mac windows() on windows x11() on linux ?Devices for list of devices available eg for pdf pdf(file = "...") ...plotting.... dev.off() ## Close the graphics device! Vector formats: pdf svg wim.metafile postscript Not so great with many individual points (file size) Resize well. http://research.stowers-institute.org/mcm/plotlayout.pdf Bitmaps: png. jpeg. lossy compression tiff. lossless compression Generally dont resize well good for many many points. Many devices -------------- Can only plot to the *active* device dev.cur() 0 returns graphics device num >=2 dev.set(<int>) change the active device dev.copy dev.copy2pdf - copy active plot to new plot E.g: ...plot... dev.copy(png, file = "filename.png") dev.off() hierachical clustering ======================= - what is close/ - how do we group things - How do we visualise the grouping - How do we interpret the grouping Agglomerative approach: - Find closest two things - requires knowing what "closest" means - continuous - euclidean sqrt( (x1-x2)^2 + (y1-y2)^2) - extends to hi D probs - Continuous - correlation similarity - Manhatten - total(x) + total(y) - Put them together - a "merge" - How do we merge? - What represents the new location - "average linkage" - ave of x and y coords - center of grav. - "complete linkage" - take furtherst two points of clusters as dist. - Repeat - A tree results shoing the ordering of how things were grouped. To simulate a simple 2D dataset: set.seed(1234) par(mar = c(0, 0, 0, 0)) x <- rnorm(12, mean = rep(1:3, each=4), sd=0.2) y<- rnorm(12, mean = rep(c(1, 2, 1), each = 4), sd = 0.2) plot(x,y, col = "blue", pch=19, cex=2) text(x+0.1, y+0.1, labels=as.character(1:12)) df <- data.frame(x = x, y = y) distxy <- dist(df) # cacls dist between all different rows/cols combos in df. hCllust <- hclust(distxy) plot(hCllust) heatmap() for looking at matrix data - does a hierachical cluster of rows and cols of the matrix rows are like observations cols are like sets of observations organises the rows and columns of the table to visualise groups or blocks within the table k-means clustering =================== K-means is NOT deterministic. If you run multiple times but resulst vary to much the alg might not be stable w.r.t to your data! It is a partitioning approach - Define #groups you would - Get "centroids" for each cluster - Assigned to closests centroid - Recalc centroids and iterate set.seed(1234) par(mar = c(0, 0, 0, 0)) x <- rnorm(12, mean = rep(1:3, each=4), sd=0.2) y <- rnorm(12, mean = rep(c(1, 2, 1), each = 4), sd = 0.2) plot(x,y, col = "blue", pch=19, cex=2) text(x+0.1, y+0.1, labels=as.character(1:12)) df <- data.frame(x = x, y = y) kmeansObj <- kmeans(df, centers = 3) par(mar = rep(0.2, 4)) plot(x, y, col = kmeansObj$cluster, pch=19, cex=2) points(kmeansObj$centers, col = 1:3, pch=3, cex=3, lwd=3) Lattice plotting system ========================= - Good for high density plots - package(lattice) - xyplot - scatter plots - bwplot - box plots - histogram - stripplot - dotplot - splom - scatter plot matrix. like pairs() in base plot - levelplot - image data - Everything (i.e. entire plot) is done in a single function call - lattice functions return calls called "trellis". this must be printed to the device to dislplay it on. Auto-prints for desktop display xyplot xplot(y ~ x |, data) y is the y data x is the x data condition vars (optional). they are factor variables and conditioning on them creates n graphs for each level of the condition var data is the dataframe eg library(datasets) library(lattice) airqual <- transform(airquality, Month=factor(Month)) xyplot(Ozone ~ Wind | Month, data = airqual, layoutc=c(5,1)) panel functions Each panel represents a subset of the data defined by the conditioning variable supplied xyplot(y ~ x | f, panel = function(x,y, ...) { panel.xyplot(x, y, ...) panel.abline(h=median(y), lty=2) }) By giving the panel function we specify how each panel is to be drawn. Each panel in this case does an xyplot, but also does a horizonal line at the data median. Note: data for each panel is different we're just saying what is to be done with the data on each panel. panel.lmline() for least regression line ggplot plotting system ======================= Grammar of Graphics by Leland Wilkinson Built on the grid graphics system. qplot() -------- Works like the plot() function of the base system data must always come from a dataframe factors are used to indicate *subsets* of the data eg qplot(x, y, data=dataframe) where x and y are column names in dataframe to highlight subgroups use qplot(x,y,data=df, color=z) where z is another column inthe dataframe to add smoother do qplot(x,y,data=df, geom=c("point", "smooth")) to do histogram qplot(x, data=df, fill=z) here fill is like color qplot(x, data=df, geom="density") for a desity smooth of hist to do panels you use facets qplot(x,y, data=df, facets=.~z) or qplot(x,y, data=df, facets=z~.) it is rows~cols and the "dot" means wildcard ggplot() --------- Plots built up in layers - plot data - overlay summary - add regressions etc Basic ggplot: g <- ggplot(dataframe, aes(x, y[, color=z])) ^^^ the aesthetics (cols to be plotted) So far there are no layers on the plot - nothing to specify how to draw the plot So we need to add this to the plot... p <- g + geom_point() ## adds "how to draw" information print(p) ## display (or use autoprinting) or use geom_line() Add a smoother g + geom_point() + geom_smooth(method="...") Add a facet grid g + geom_point + facet_grid(. ~ z) + .... ^^^^^^ works the same as qplot To annotate the plots use xlab(), ylab(), labs(), ggtitle() eg labs(title="...", x="...", y="...") theme() theme_gray(), theme_bw() some standard appearances Aesthetics options - color=... - size=... - alpha=.. The smoother. - size= - linetype= - method=.. - se=... Note: ylim() will FILTER the data to filter out things outside limits. To keep outliers use coord_cartesian(ylim=()) Categorise on continuos ver: To categorize continuous variables use CUT() function! Can use quantile() function to get some reasonable cut points